Research
Our overarching goal is to explore and understand how phenotype emerges from the interactions of the individual constituents of the living systems.
We instantiate this question to three particular areas of interest:
Drug interactions. The relative yearly gain of new medicines available for treating common diseases is in general decreasing, but the overall absolute number of bioactive compounds is staggering. There is a large potential in combining existing treatments to increase their effectivness. However, the combinatorial explosion is preventing the testing of all possible combinations for each purpose ‒ for n drugs there is on the order of n2 pairwise combinations! There is clearly a need for a predictive model that would enable us to prioritise the testing of particular combinations.
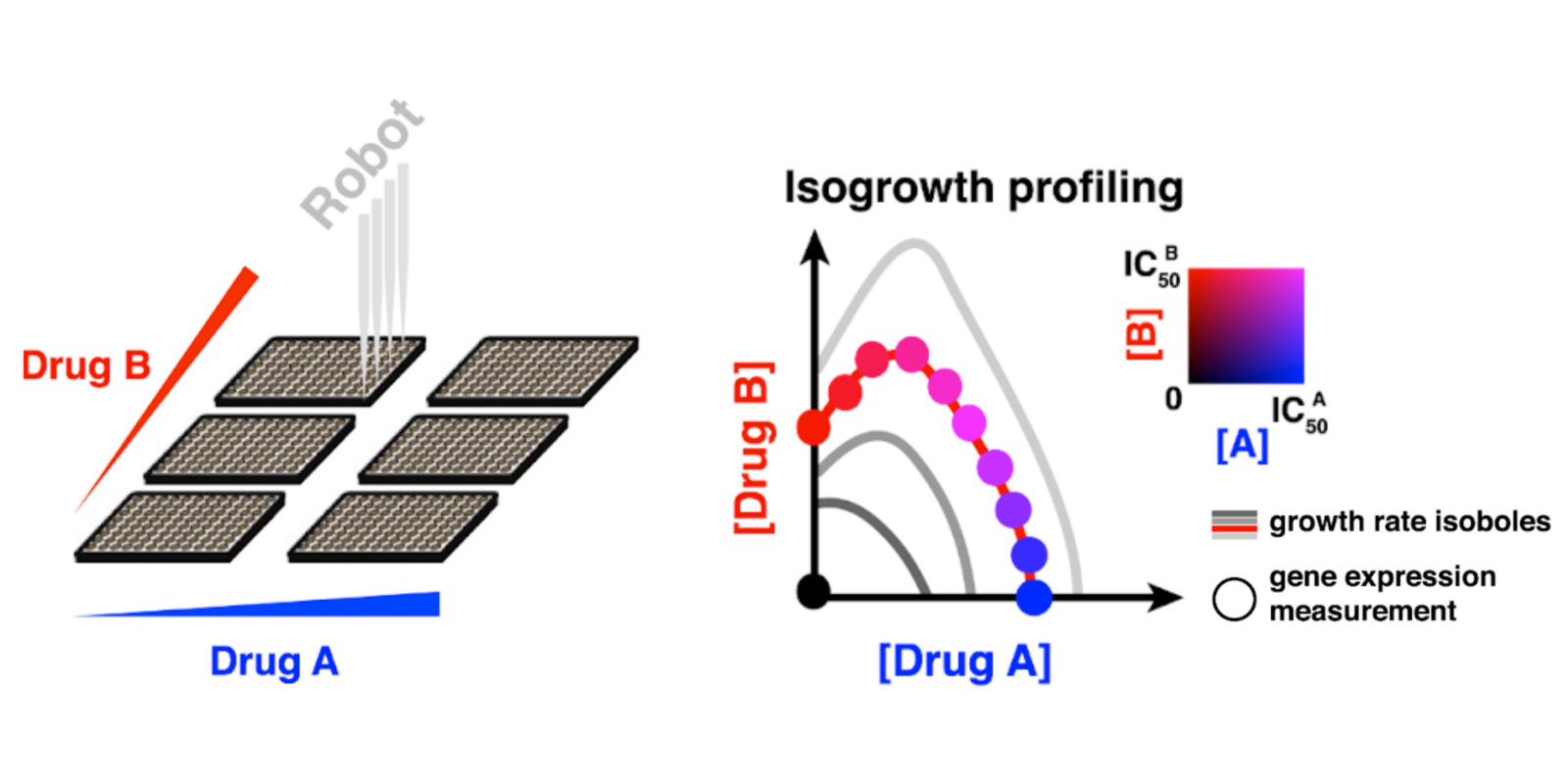 Our working hypothesis is that one of the major factors preventing the prediction of drug interactions is the confusion of gene expression responses to the drug with the gene expression changes due to altered growth rate. In addition, different drugs take different amount of time to reach the steady state effect, but this is rarely reflected in experimental protocols measuring the gene expression response. We have developed an assay to experimentally side-line both these problems, which we term isogrowth profiling. We measure gene expression changes in various concentrations of two drugs such that the resulting steady state inhibition is constant. This allows us to disentangle the physiological effects of individual drugs, predict hihger-order drug interactions as well as pair-wise drug interactions from individual drug gene expression responses. While we have been able demonstrate this with a handful of drugs, we are now working on a simplified version of isogrowth profiling that would enable us to scale up this effort and really show the utility of isogrowth profiling for drug interaction prediction.
Our working hypothesis is that one of the major factors preventing the prediction of drug interactions is the confusion of gene expression responses to the drug with the gene expression changes due to altered growth rate. In addition, different drugs take different amount of time to reach the steady state effect, but this is rarely reflected in experimental protocols measuring the gene expression response. We have developed an assay to experimentally side-line both these problems, which we term isogrowth profiling. We measure gene expression changes in various concentrations of two drugs such that the resulting steady state inhibition is constant. This allows us to disentangle the physiological effects of individual drugs, predict hihger-order drug interactions as well as pair-wise drug interactions from individual drug gene expression responses. While we have been able demonstrate this with a handful of drugs, we are now working on a simplified version of isogrowth profiling that would enable us to scale up this effort and really show the utility of isogrowth profiling for drug interaction prediction.
Interactions of growth sensitive cellular components.
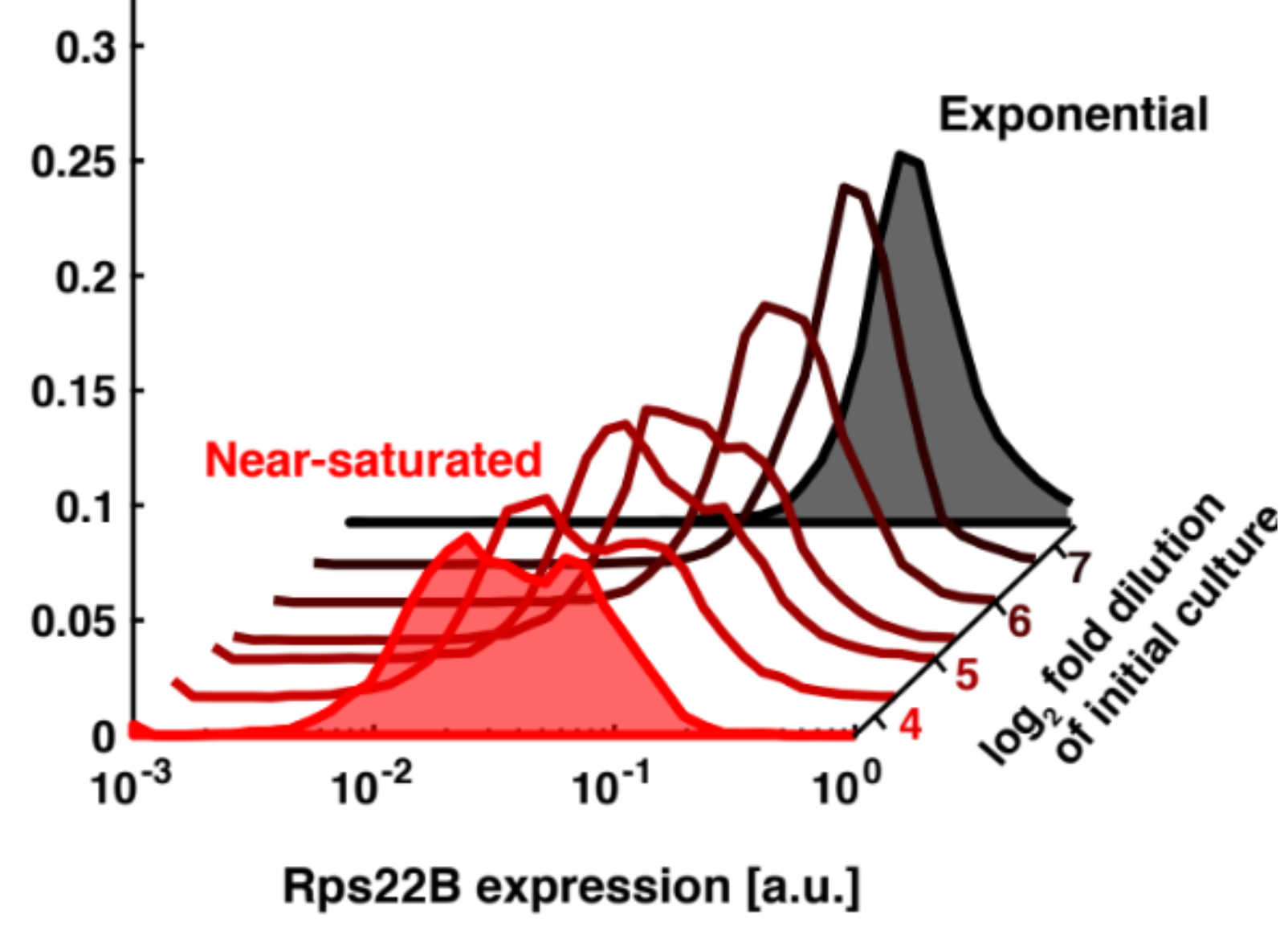 Isogrowth profiling, while developed for predicting drug interaction, at the same time happens to be of great utility when studying processes that are highly dependent on growth rate. Perhaps the most importnat such process is translation and the regulation thereof. Traditionally, many cellular processes and their regulation has been studied using perturbation with small molecule drugs. However, for given dosage of the drug, a plethora of factors in the growth medium influence the resulting growth rate. When using these inhibitors to study translation, regulatory process that manifest only at certain growth rates might elude us if in fact it is not possilbe to achieve such growth rate using the perturbagen itself. To circumvent this, we use isogrowth profiling, where we supply another drug to achieve a variety of growth rates with the same dosage of the perturbagen of interest present. In this way, we have succeeded in uncovering a completely new role for intronic sequences in the genes coding for ribosomal proteins in the budding yeast - the diversification of the population in face of looming nutrient starvation. Our aim is to apply this approach to study further regulation of not only translation, but also other processes greatly dependent on growth rate - such as quorum sensing or respiration.
Isogrowth profiling, while developed for predicting drug interaction, at the same time happens to be of great utility when studying processes that are highly dependent on growth rate. Perhaps the most importnat such process is translation and the regulation thereof. Traditionally, many cellular processes and their regulation has been studied using perturbation with small molecule drugs. However, for given dosage of the drug, a plethora of factors in the growth medium influence the resulting growth rate. When using these inhibitors to study translation, regulatory process that manifest only at certain growth rates might elude us if in fact it is not possilbe to achieve such growth rate using the perturbagen itself. To circumvent this, we use isogrowth profiling, where we supply another drug to achieve a variety of growth rates with the same dosage of the perturbagen of interest present. In this way, we have succeeded in uncovering a completely new role for intronic sequences in the genes coding for ribosomal proteins in the budding yeast - the diversification of the population in face of looming nutrient starvation. Our aim is to apply this approach to study further regulation of not only translation, but also other processes greatly dependent on growth rate - such as quorum sensing or respiration.
Interactions of components made faulty by ageing. Some diseases have a single, strong perturbagen as their cause. Some others consist of plethora tiny perturbations that accumulate and together interact to perturb the living system, leading to its detriment. A perfect example of the latter is ageing, making it an excellent use case to apply our knowledge of life as an interacting network towards imporvement of human health. We are involved in a number of computational projects aimed at characterising the network changes during human ageing, specifically in the immune system, to disentangle their drivers and interactions thereof that bring about the frailty and multi-morbidity currently associated with old age.